The Unique Problems of Medical Computer Vision
You have never seen such an exhausted disdain as when you ask an overworked radiologist to segment brain lesions on an additional thousand MR scans….

You have never seen such an exhausted disdain as when you ask an overworked radiologist to segment brain lesions on an additional thousand MR scans. Embarrassing to say now, but this was once me slowly burning the goodwill of my neuroradiologist collaborators for my thesis project. Coming from a computer science background where our datasets comprise tens of thousands of images at a minimum, it would seem insane to embark on a project with only a hundred patients’ worth of data. But the truth is, computer vision and AI scientists often forget that there’s a very real cost for annotating all of this data.
Do no harm.
We all know that annotations for radiology scans are more expensive to get than equivalent labels for cats and dogs on images. For most medical tasks, we can’t crowdsource our annotation process to something like Amazon Mechanical Turk. The result is that this cost is not just monetary, but is an additional burden on our healthcare system and the thinly-stretched experts that have to leverage their expertise to create the data labels. I was confronted with this fact when one of my exasperated radiology colleagues finally asked me: “At what point does starting a medical AI project cross the line into healthcare burden? At what point are you doing harm?” It took me a hot minute to accept the premise of the question, but since then, it’s been one of the most foundational pillars in how I personally approach computer vision for medical images. Every label we ask a doctor to make is a patient that they could be diagnosing instead.

So, does that mean we should give up? Is medical computer vision at a strict disadvantage to other vision fields because of the lack of labeled data? Well, not exactly. Let’s take a look at the “mean image” of two datasets: (a) the famous computer vision dataset of ImageNet and (b) the CheXpert dataset from Stanford. These images were created by taking the mean value of every pixel of every image at every different location. The mean ImageNet image just looks like a blur, caused by the extremely heterogeneous images that comprise ImageNet. However, the mean image of all the chest X-Rays still retains much of the chest X-Ray characteristics. This is due to medical images having less inter-image variance; two chest X-Rays images look much more similar to each other than two different dog images from ImageNet. However, the signal we’re looking for (e.g. Normal CXR vs. pneumonia) is also much smaller than the difference between a cat image and a dog image from ImageNet. This is the very unique problem of medical computer vision: we are attempting to solve a small signal on the background of small noise whereas standard computer vision’s problem is a large signal on the background of large noise. I think we often forget that medical images are not the same as the images you’ll find on your smartphone. The field of computer vision and AI often forgets how beautiful and unique the problem of medical imaging can be.
When you have a hammer, everything’s a nail.
And boy howdy, is deep learning a hammer. As tempting as it is to chock a problem up to collecting more data, we want to push a different approach at Sirona: we want to invent new tools, one tailored to the specific problem of medical images. We want to take advantage of the idea that you don’t need that many images to explore the manifold for most medical image modalities. The focus should be on determining how to robustly learn the intricate differences between clinically relevant contexts given to us by the medical images.
Sirona’s unified platform and strong network of radiology partners is the perfect warehouse to develop that new toolkit. Rather than rely solely on a supervised approach, we want to look at techniques such as metric learning, weakly-/semi-supervised learning, and human-in-the-loop training to ensure that we can be more efficient with our training data. By combining a small corpus of expert labels with more natural language processing (NLP) derived labels from reports, we can train our networks on all the data within our system without having to pass the burden of image annotation onto the very radiologists we aim to serve.
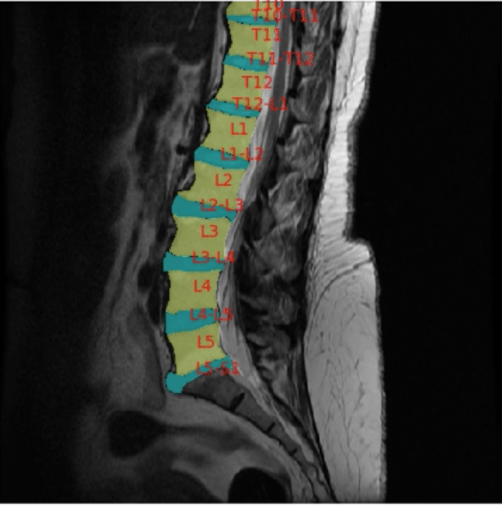
But having the right hammer is only half the story; we need to make sure we’re hitting the right nail. The most innovative piece of AI capability is meaningless if it can’t be leveraged to improve the healthcare system. Let’s be honest, basic anatomical segmentation of spine MR’s is not the sexiest of medical AI projects currently being done. But seeing those network outputs be integrated into the Anatomic Navigator feature and seeing the positive impact on radiology workflow is a clear signal in what computer vision could do for our healthcare system.
I’d like to think I’ve grown since my early grad school days when I treated my clinician partners like labelers. I now understand the value added by our radiologists are not the annotations they provide, but the clinical guidance they give in how we build our algorithms. To me, the unique problem of medical imaging isn’t in signal to noise ratios or figuring out how to solve the small data problem. It’s in understanding how to build meaningful applications that encapsulates clinical knowledge to truly make a difference in the world.
Darvin Yi is part of the Sirona R&D team and works on numerous projects involving computer vision and AI algorithm development. Some of the product ideas and features described in this article are future oriented and may not be reflected in Sirona’s commercial applications.
MKT018 Rev A.0
